Single-Cell Analysis Links Immune Cells to Kidney Cancer Recurrence
The immune nature of renal carcinoma stands out when compared to other cancers: More immune cells infiltrate kidney cancers than most other solid tumors, and kidney cancer is one of the most responsive malignancies to today’s immunotherapy regimens. But despite treatment, about 40 percent of patients with resected clear cell renal carcinoma will relapse and develop metastatic disease; the five-year survival in 10 percent of patients underscores the need to understand the cellular and molecular mechanisms in primary lesions that are predictive for recurrence.
In the May 2021 issue of Cell, Columbia University researchers published the results of their study showing that the presence of a rare and previously unknown type of immune cell in kidney tumors can predict which patients are likely to have cancer recur after surgery. These cells could even be driving aggressive disease.
“The presence of these cells provides us with the potential to identify patients at high risk of disease recurrence after surgery and therefore may be candidates for more aggressive therapy,” says co-senior author James M. McKiernan, MD, Urologist-in-Chief, NewYork-Presbyterian/Columbia University Irving Medical Center. Andrea Califano, Dr, Chair, Department of Systems Biology at Columbia University Vagelos College of Physicians and Surgeons, also served as co-senior author.
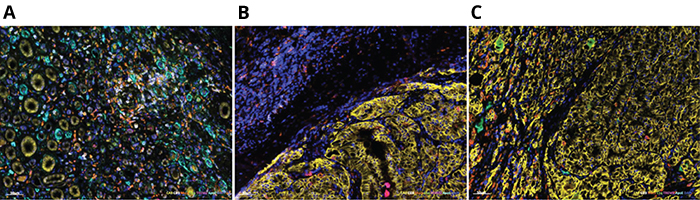
(A) Adjacent normal kidney tissue; (B) Tumor stroma; (C) Tumor (Images courtesy of Aleksandar Obradovic)
Though kidney tumors are densely infiltrated by immune cells, cell types and their association with post-surgical outcomes have remained incompletely characterized. To uncover the fine details of the immune cells that infiltrate kidney cancers, the researchers used single-cell RNA sequencing, a high-throughput technique that enables investigators to obtain snapshots inside tens of thousands of cells from one tumor in a single experiment, providing insights into the identity and behavior of the various cell types. Because single-cell sequencing works by detecting a small number of mRNA molecules inside each cell, it often fails to detect the mRNAs of genes with low expression levels – a phenomenon known as gene dropout – including key signaling genes and drug targets such as immunotherapy checkpoints.
Prediction Algorithm Addresses Gene Dropout
The researchers addressed gene dropout by developing a prediction algorithm – metaVIPER, which builds on the VIPER algorithm developed in the Califano Laboratory. The new algorithm can infer which genes are active by looking at the expression of other related genes. With the addition of metaVIPER, the Columbia team estimates they can accurately detect the activity of 70 to 80 percent of all regulatory genes in each cell, eliminating dropout across cells.
The researchers used this combined approach to analyze more than 200,000 tumor cells and normal cells in adjacent tissue taken from 11 patients with clear cell renal carcinoma who underwent surgery by Columbia urologists. The analysis revealed a unique subpopulation of macrophages – C1Q+ – found only in tumors and associated with eventual relapse of disease after initial treatment. The VIPER analysis also revealed the master regulators that control the activity of these macrophages.
This “signature” was validated in a second set of patient data obtained through collaboration with researchers from Vanderbilt University who showed that the signature strongly predicted relapse in a second set of over 150 patients.
Furthermore, these macrophages were found to interact directly with tumor cells through receptor-ligand gene pairs. “These data raise the intriguing possibility that these macrophages are not just markers of more risky disease but may actually cause the disease to recur and progress and that targeting these cells could improve clinical outcomes,” notes Aleksandar Obradovic, an MD/PhD student at Columbia and the study’s co-first author.
“VIPER-based technologies, such as the Oncotreat test, could be used to identify drugs targeting these rare but critical subpopulations, thus preventing the poor outcomes associated with their presence,” says Dr. Califano.
The combination of single-cell sequencing with the VIPER algorithm has the potential to dissect other types of cancer as well, the researchers say. The two techniques, when combined, are highly effective at characterizing the cells within a tumor and in surrounding tissues and should have broad applicability, even beyond the study of cancer.
Read More
Single-cell protein activity analysis identifies recurrence-associated renal tumor macrophages. Obradovic A, Chowdhury N, Haake SM, Ager C, Wang V, Vlahos L, Guo XV, Aggen DH, Rathmell WK, Jonasch E, Johnson JE, Roth M, Beckermann KE, Rini BI, McKiernan J, Califano A, Drake CG. Cell. 2021 May 27;184(11):2988-3005.e16.